|
|
@@ -1,9 +1,137 @@
|
|
|
-# Deploying
|
|
|
+# Hartman Lab Server Manual
|
|
|
|
|
|
-Server scripts and configs are organized using [GNU Stow](https://github.com/aspiers/stow) packages.
|
|
|
+## First-time login
|
|
|
|
|
|
-Examples:
|
|
|
+1. Ensure an admin has enabled your user account and provided a username.
|
|
|
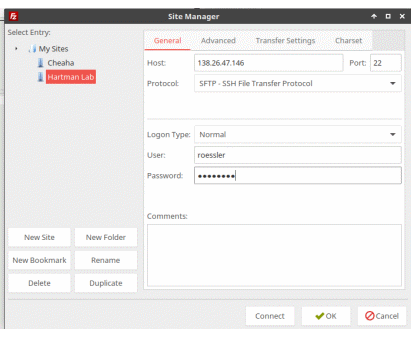
+2. Login via [`ssh`](#clients): `ssh username@hartmanlab.genetics.uab.edu` (the default password is your username)
|
|
|
+3. You will be prompted to create a new password and automatically logged out
|
|
|
+4. Login again using new password: `ssh username@hartmanlab.genetics.uab.edu`
|
|
|
+5. Change `samba` password (default password is also your *username*): `smbpasswd`
|
|
|
+6. To change your password again in the future: `passwd`
|
|
|
+7. *Optional*: Generate a public-private keypair on your client and copy it to the server for faster and more secure logins.
|
|
|
|
|
|
-* Deploy system-wide MATE layout and themes: `sudo stow --adopt -R -t / theme`
|
|
|
-* Deploy system-wide scripts: `sudo stow --adopt -R -t / scripts`
|
|
|
-* Deploy system config files: `sudo stow --adopt -R -t / config`
|
|
|
+ ```bash
|
|
|
+ ssh-keygen -t rsa -b 4096 -f ~/.ssh/id_rsa_4096
|
|
|
+ ssh-copy-id -i ~/.ssh/id_rsa_4096.pub username@hartmanlab.genetics.uab.edu
|
|
|
+ ```
|
|
|
+
|
|
|
+ Users may need to add their keypair to PuTTy/Filezilla/WinSCP manually.
|
|
|
+
|
|
|
+## `ssh`
|
|
|
+
|
|
|
+Connect to the server remotely using the command line.
|
|
|
+
|
|
|
+* Linux/OSX
|
|
|
+ * `ssh username@hartmanlab.genetics.uab.edu`
|
|
|
+* Windows
|
|
|
+ * [PuTTY](http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html)
|
|
|
+* Android
|
|
|
+ * JuiceSSH
|
|
|
+ * Termux
|
|
|
+
|
|
|
+### X forwarding
|
|
|
+
|
|
|
+Launch graphical server programs locally on a client that run on the server.
|
|
|
+
|
|
|
+* Linux/OSX
|
|
|
+ * Enable X forwarding during ssh login: `ssh -X username@hartmanlab.genetics.uab.edu`
|
|
|
+* Windows
|
|
|
+ * Install [Xming](http://www.straightrunning.com/XmingNotes/) and enable X11 forwarding in the [PuTTY](http://www.chiark.greenend.org.uk/~sgtatham/putty/download.html) options.
|
|
|
+
|
|
|
+## `sftp`
|
|
|
+
|
|
|
+Browse and manage files stored on the server.
|
|
|
+
|
|
|
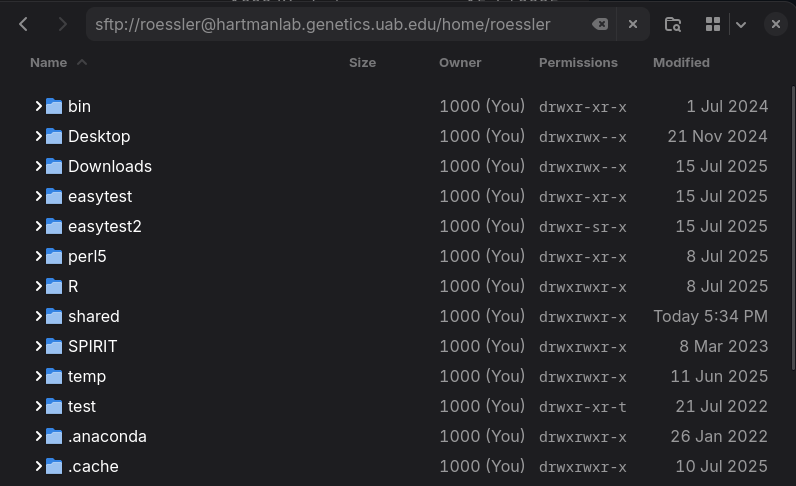
+* Native access to sftp shares using client file manager (`sftp://username@hartmanlab.genetics.uab.edu:/home/username`):
|
|
|
+ 
|
|
|
+* Linux/OSX/Windows
|
|
|
+ * [Filezilla](https://filezilla-project.org/download.php?type=client)
|
|
|
+
|
|
|
+ 
|
|
|
+ * [sshfs](https://www.digitalocean.com/community/tutorials/how-to-use-sshfs-to-mount-remote-file-systems-over-ssh)
|
|
|
+* Windows
|
|
|
+ * [WinSCP](https://winscp.net/eng/index.php)
|
|
|
+
|
|
|
+## `samba`
|
|
|
+
|
|
|
+Samba file shares can be mounted cross-platform as if th server data existed locally. The server provides two shares:
|
|
|
+
|
|
|
+1. Shared data array (`/mnt/data`): `\\username\\data`
|
|
|
+2. User's `$HOME` directory: `\\username\\username`
|
|
|
+
|
|
|
+The default `samba` credentials are the same as your server username and password. Users can change their `samba` password using `smbpasswd`.
|
|
|
+
|
|
|
+## `x2goclient`
|
|
|
+
|
|
|
+Users can access an X2Go remote desktop session using `x2goclient` from a client. `x2goclient` is available for Linux/OSX/Windows from the [X2Go website](http://wiki.x2go.org/doku.php) or by installing the `x2goclient` package.
|
|
|
+
|
|
|
+X2Go sessions can be paused or suspended from the X2Go client window. Multiple sessions can be created on the client, making it possible to select alternate quality settings based on location and bandwidth.
|
|
|
+
|
|
|
+**Note:** Some programs do not continue to run at full speed when an X2Go session is paused. In these cases, the program should be run via `ssh` in a [`tmux`](https://en.wikipedia.org/wiki/Tmux) or [`screen`](https://www.gnu.org/software/screen/) session.
|
|
|
+
|
|
|
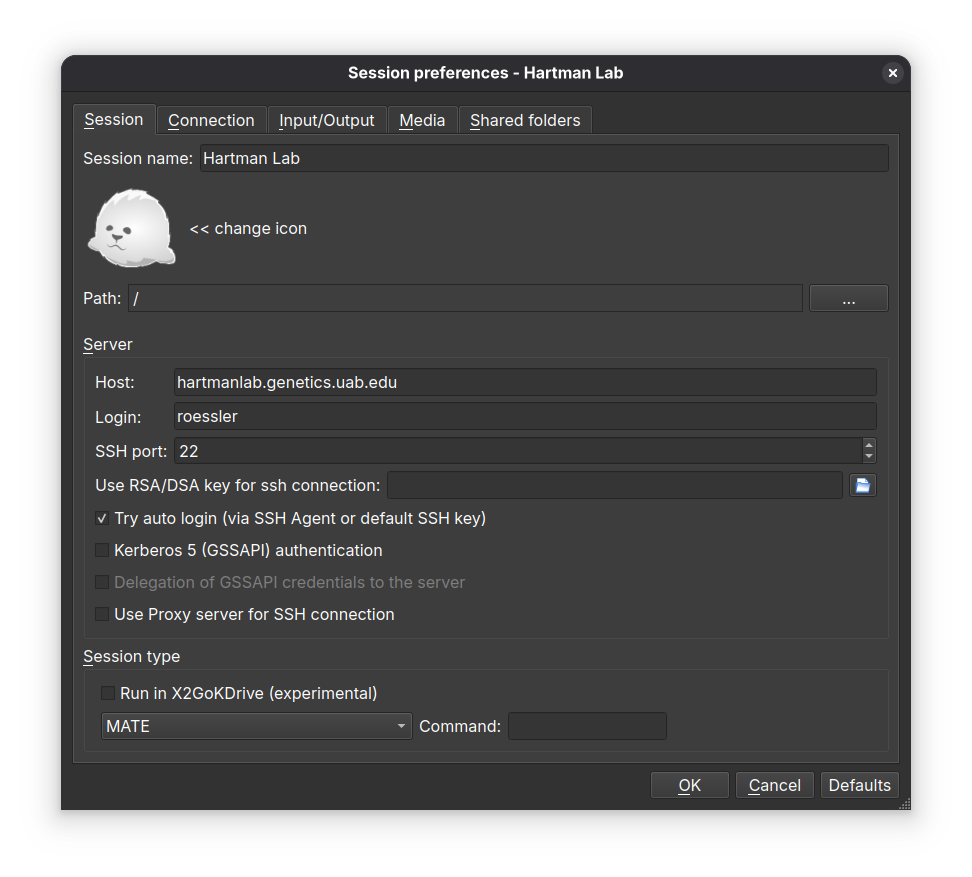
+* Session tab
|
|
|
+ * Session name: Hartman Lab Server
|
|
|
+ * Host: `hartmanlab.genetics.uab.edu`
|
|
|
+ * Login: *`username`*
|
|
|
+ * SSH port: `22`
|
|
|
+ * Session type: **MATE** (MATE provides the best experience with X2Go)
|
|
|
+ 
|
|
|
+* Connection tab
|
|
|
+ * Set the connection speed to LAN when connecting from within the UAB network and WAN when outside of the UAB network
|
|
|
+ * Compression settings should be left unchanged or set to *adaptive*.
|
|
|
+* Input/output tab
|
|
|
+ * If automatic window resizing is not working properly (common on HiDPI monitors), set the desired startup window resolution size manually. For full screen sessions, this should match your client display. In case of scaling issues, play with the DPI setting, 96 is a sane starting value.
|
|
|
+ * If there are any issues with keyboard mapping (ex. the arrow keys are not working), select *Configure Keyboard* and leave the default selected settings.
|
|
|
+* Media tab
|
|
|
+ * Disable sound support. This will prevent pulseaudio from spamming the server logs.
|
|
|
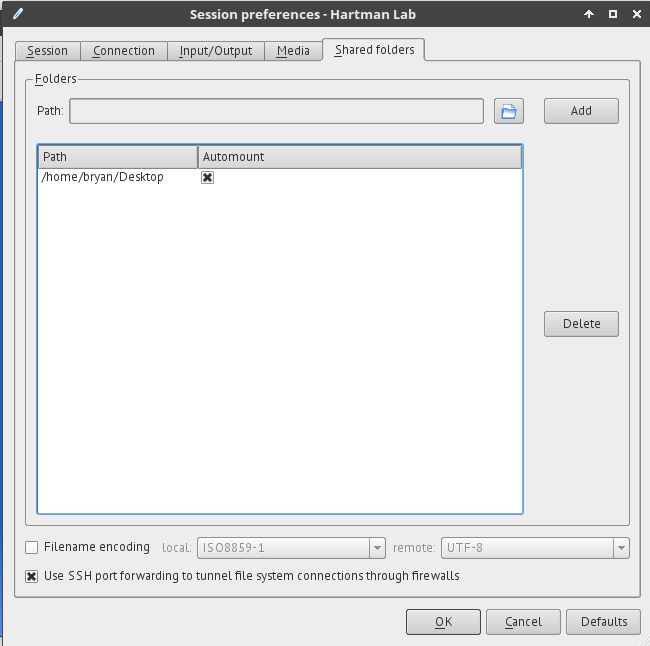
+* Shared folders tab
|
|
|
+ * Select folders on the client to be shared with the server during a session. Browse to the chosen folder, add it to the share, and select *automount*.
|
|
|
+ * These folders will then appear on the server under `/media/disk/<share_name>`.
|
|
|
+ * Shared folder permissions can be modified by admin for multi-user access.
|
|
|
+ 
|
|
|
+
|
|
|
+## Robot computer remote desktop access
|
|
|
+
|
|
|
+In an X2Go session, go to *Applications>Internet>Remote Viewer>Connection>New* and enter `vnc://192.168.16.101:5900`
|
|
|
+
|
|
|
+## Webcam robot monitoring
|
|
|
+
|
|
|
+* In an X2Go session, via a web browser at [`http://localhost:9999`](http://localhost:9999)
|
|
|
+* Locally via a web browser via an SSH tunnel: `ssh -f username@hartmanlab.genetics.uab.edu -L 9999:localhost:9999 -N`
|
|
|
+
|
|
|
+## RStudio Server
|
|
|
+
|
|
|
+* In an X2Go session, via a web browser at [`http://localhost:8787`](http://localhost:8787)
|
|
|
+* Locally via a web browser via an SSH tunnel: `ssh -f username@hartmanlab.genetics.uab.edu -L 8787:localhost:8787 -N`
|
|
|
+
|
|
|
+## JupyterLab (Python) IDE
|
|
|
+
|
|
|
+* In an X2Go session, via a web browser at [`http://localhost:8888`](http://localhost:8888)
|
|
|
+* Locally via a web browser via an SSH tunnel: `ssh -f username@hartmanlab.genetics.uab.edu -L 8888:localhost:8888 -N`
|
|
|
+
|
|
|
+## Data backup
|
|
|
+
|
|
|
+[`rsync`](https://linux.die.net/man/1/rsync) is recommended for users that would just like to periodically backup their $HOME directory to a local machine.
|
|
|
+
|
|
|
+* Copy a user's `$HOME`' directory locally to `/home-backup`: `rsync -azH --delete username@hartmanlab.genetics.uab.edu:/home/username/ home-backup/`
|
|
|
+* Copy a shared directory locally `rsync -azh username@hartmanlab.genetics.uab.edu:/mnt/data/scans/20250723_roessler_project .`
|
|
|
+
|
|
|
+## Resources
|
|
|
+
|
|
|
+* [RHEL documentation](https://access.redhat.com/documentation/en/red-hat-enterprise-linux/)
|
|
|
+* [Navigating the Linux CLI](https://www.digitalocean.com/community/tutorials/basic-linux-navigation-and-file-management)
|
|
|
+* [Explainshell](https://explainshell.com/)
|
|
|
+* [UAB Cheaha](https://docs.uabgrid.uab.edu/wiki/Cheaha_GettingStarted)
|
|
|
+
|
|
|
+## Platform
|
|
|
+
|
|
|
+* AlmaLinux 9.6 w/ Linux 6.1 LTS Hyperscale SIG kernel
|
|
|
+* Intel Xeon X99 E5-2650v4 CPU
|
|
|
+* 96GB RAM
|
|
|
+* 4TB PCIE3 NVMe SSD: `/`, `/home`
|
|
|
+* 20TB btrfs raid1 array: `/mnt/data`
|
|
|
+* 20TB btrfs raid1 backup array: `/mnt/backup`
|
|
|
+
|
|
|
+## Administrators
|
|
|
+
|
|
|
+See additional documentation in [`docs/README.md`](docs/README.md)
|